Quick Start
The objective of this tutorial is to reproduce part of the results presented in Baurens et al (2019) and Ahmed et al (2019), using respectively VCFHunter and TraceAncestor.
The outputs of these programs can then be used in the GeMo webapps.
Installation requirements
This tutorial is developed to run on Linux or Apple (MAC OS X) operating systems. There are no versions planned for Windows.
Software requirements:
Perl 5 for TraceAncestor
Python 3 for VCFHunter
Testing your Perl installation
To test that Perl 5 is installed, enter on the command line
perl -version
Testing your Python installation
To test that Python 3 is installed, enter on the command line
python3 --version
Now, you can clone the repository, create a virtualenv and install several additionnal package using pip.
git clone https://github.com/gdroc/GeMo_tutorials.git
cd GeMo_tutorials
python3 -m venv $PWD/venv
source venv/bin/activate
pip install numpy
pip install matplotlib
pip install scipy
Download Dataset
For this tutorial, Dataset that will be used by TraceAncestor or by VCFHunter are accessible on Zenodo https://doi.org/10.5281/zenodo.6539270
To download this, you only need to launch the script download_dataset.pl without any parameter
perl download_dataset.pl
This script create a new directory data
data/
├── Ahmed_et_al_2019_color.txt
├── Ahmed_et_al_2019_individuals.txt
├── Ahmed_et_al_2019_origin.txt
├── Ahmed_et_al_2019.vcf
├── Baurens_et_al_2019_color.txt
├── Baurens_et_al_2019_individuals.txt
├── Baurens_et_al_2019_origin.txt
├── Baurens_et_al_2019_chromosome.txt
└── Baurens_et_al_2019.vcf
These files are require for this tutorial to run VCFHunter or TraceAncestor
Input
Baurens_et_al_2019_origin.txt : A two column file with individuals in the first column and group tag (i.e. origin) in the second column
individuals |
origin |
|---|---|
P2 |
AA |
T01 |
BB |
T02 |
BB |
T03 |
AA |
T04 |
AA |
T05 |
AA |
T06 |
AA |
T07 |
AA |
T08 |
BB |
Baurens_et_al_2019.vcf : A vcf file with ancestral and admixed individuals
grep #CHROM data/Baurens_et_al_2019.vcf
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT ACC48-FPG ACC48-FPN ACC48-P_Ceylan ACC48-Red_Yade DYN163-Kunnan DYN275-Pelipita DYN359-Safet_Velchi GP1 GP2 P1 P2 T01 T02 T03 T04 T05 T06 T07 T08 T10 T11
Baurens_et_al_2019_individuals.txt : A two column file with individuals to scan for origin (same as defined in the VCF headerline) in the first column and the ploidy in the second column.
Baurens_et_al_2019_color.txt : A color file with 4 columns: col1=group and the three last column corresponded to RGB code.
group |
name |
r |
g |
b |
|---|---|---|---|---|
AA |
acuminata |
0 |
255 |
0 |
BB |
balbisiana |
255 |
0 |
0 |
Run workflow using create_gemo_input.pl
perl create_gemo_input.pl --help
Parameters :
-v, --vcf A vcf file [required]
-o, --origin A two column file with individuals in the first column and group tag (i.e. origin) in the second column [Required]
-i, --individuals List of individuals to scan from vcf, as defined in the VCF headerline [Required]
-m, --method Permissible values: vcfhunter traceancestor (String). Default vcfhunter
-c, --color A color file with 4 columns: col1=group and the three last column corresponded to RGB code.
-t, --threads Number of threads
-d, --dirout Path to the output directory (Default method option name)
-h, --help display this help
1. With VCFHunter method
You must use the dataset prefixed with Baurens_et_al.
perl create_gemo_input.pl --vcf data/Baurens_et_al_2019.vcf --origin data/Baurens_et_al_2019_origin.txt --individuals data/Baurens_et_al_2019_individuals.txt --method vcfhunter --color data/Baurens_et_al_2019_color.txt --threads 4
2. With TraceAncestor method
You must use the dataset prefixed with with Ahmed_et_al.
perl create_gemo_input.pl --vcf data/Ahmed_et_al_2019.vcf --origin data/Ahmed_et_al_2019_origin.txt --individuals data/Ahmed_et_al_2019_individuals.txt --method traceancestor --color data/Ahmed_et_al_2019_color.txt
Explanation of outputs
A directory was create depending on parameter dirout (default method name)
For example, for VCFHunter, for each individual present in the file data/Baurens_et_al_2019_individuals.txt, 4 outputs are produced in this directory, prefixed with the name of indivual :
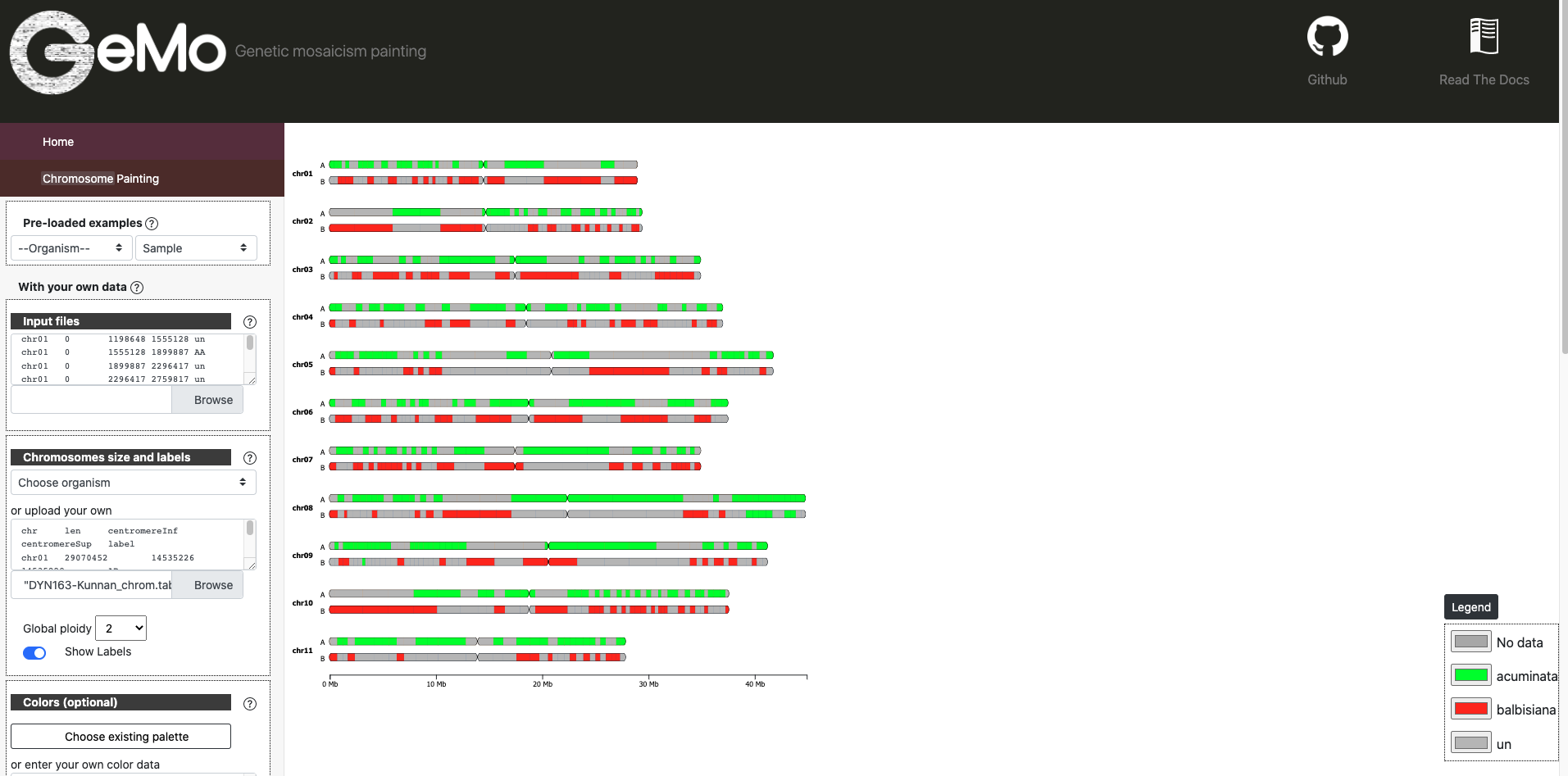
DYN163-Kunnan_ideo.txt : A text file of the position of genomic blocks the ancestry mosaic with a succession of genomic blocks along the chromosome
chr |
haplotype |
start |
end |
ancestral_group |
|---|---|---|---|---|
chr01 |
0 |
0 |
20888 |
AA |
chr01 |
0 |
20888 |
451633 |
AA |
chr01 |
0 |
451633 |
848109 |
AA |
chr01 |
0 |
848109 |
1198648 |
AA |
chr01 |
0 |
1198648 |
1555128 |
un |
chr01 |
0 |
1555128 |
1899887 |
AA |
chr01 |
0 |
1899887 |
2296417 |
un |
chr01 |
0 |
2296417 |
2759817 |
un |
DYN163-Kunnan_chrom.txt : A tab file with name, length and karyotype based on ploidy (optionaly the location of centromere).
chr |
len |
centromereInf |
centromereSup |
label |
|---|---|---|---|---|
chr01 |
29070452 |
14535226 |
14535228 |
AB |
chr02 |
29511734 |
14755867 |
14755869 |
AB |
chr03 |
35020413 |
17510206 |
17510208 |
AB |
chr04 |
37105743 |
18552871 |
18552873 |
AB |
chr05 |
41853232 |
20926616 |
20926618 |
AB |
chr06 |
37593364 |
18796682 |
18796684 |
AB |
chr07 |
35028021 |
17514010 |
17514012 |
AB |
chr08 |
44889171 |
22444585 |
22444587 |
AB |
chr09 |
41306725 |
20653362 |
20653364 |
AB |
chr10 |
37674811 |
18837405 |
18837407 |
AB |
chr11 |
27954350 |
13977175 |
13977177 |
AB |
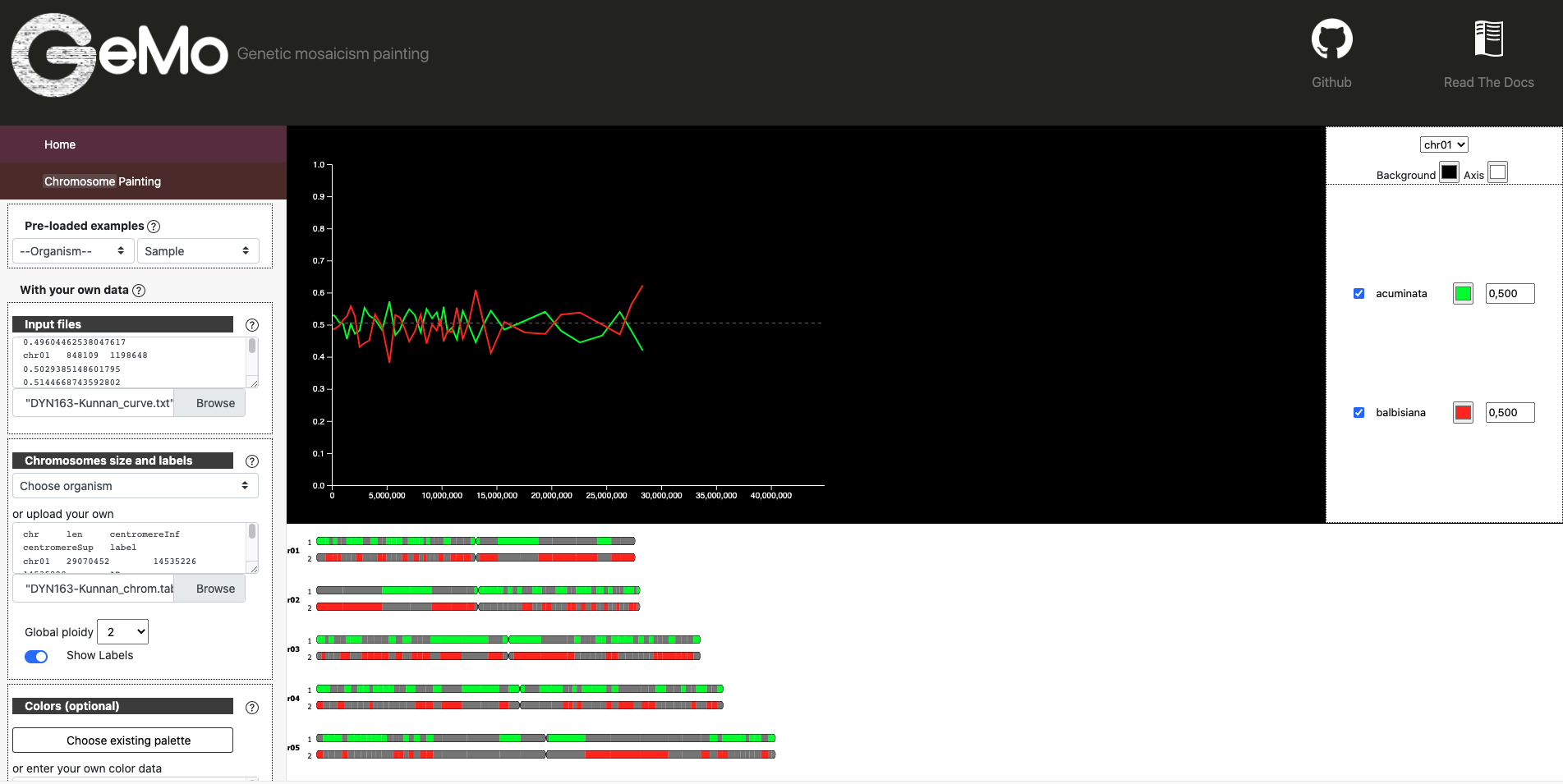
BDYN163-Kunnan_color.txt : Frequency of ancestors alleles along chromosome for the particular hybrid focused.
group |
name |
hex |
|---|---|---|
AA |
acuminata |
#00ff00 |
BB |
balbisiana |
#ff0000 |
un |
un |
#bdbdbd |
DYN163-Kunnan_curve.txt : Frequency of ancestors alleles along chromosome for the GeMo visualization tool.
chr |
start |
end |
AA |
BB |
|---|---|---|---|---|
chr01 |
20888 |
525207 |
0.660757486645395 |
0.30378982223766354 |
chr01 |
525207 |
1086954 |
0.6425583592191819 |
0.3508607451997505 |
chr01 |
1086954 |
1563263 |
0.7355412887547506 |
0.2661255866893344 |
chr01 |
1563263 |
2058335 |
0.6136974042002844 |
0.3851682528896984 |
chr01 |
2058335 |
2638987 |
0.5543371247412866 |
0.39469329280411 |
chr01 |
2638987 |
3190388 |
0.6752108036341729 |
0.3208947817296506 |
chr01 |
3190388 |
3905155 |
0.6951554613138214 |
0.3155181655339866 |
chr01 |
3905155 |
4800522 |
0.6813746934348566 |
0.32271710110143237 |
Visualization and block refinement with GeMo
Go to GeMo WebApp
Ideogram Mode
Curve mode